Molecular Genetics Panel Test Information
The team uses lab developed, targeted, and reliable Next Generation Sequencing (NGS) chemistry to provide rule out testing for all genes in all of the gene panel tests at the sensitivity of Sanger sequencing and MLPA combined, including exon-level copy number assessment for all genes.
Molecular Genetics NGS testing menu includes:
- Hereditary Cancer Panels
- Charcot-Marie-Tooth Gene Panel
- Mitochondrial Genome Sequencing
- Epilepsy Panels
- Lysosomal Storage Disorders
- Urea Cycle Disorders
- Familial Dyslipidemia/Hypercholesterolemia Panels
- Adult Haemolytic Uremic Syndrome panel (available for research purposes only)
- Hematologic Oncology Molecular Testing
- Targeted Gene Testing
The team is pleased to offer these services at competitive turn-around times (routine 4-6 weeks), at the industry leading cost efficiency.
“My clinic routinely uses LHSC molecular genetics laboratory for Charcot-Marie-Tooth Panel and Mitochondrial Genome sequencing which we previously obtained at various international laboratories. The service quality, responsiveness, reporting, and TAT meet and exceed international standards of quality at our own Canadian academic health care lab.” — Mark Tarnopolsky, MD, PhD, FRCP(C), Professor of Pediatrics and Medicine, President and CEO, Exerkine Corporation, Director of Neuromuscular and Neurometabolic Clinic, McMaster University Medical Center
Hereditary Cancer Panels (HCP)
Test Description:
The Hereditary Cancer Panel (HCP) is a genetic test designed to help assess the cancer predisposition risk for a number of common heritable cancers including: breast, ovarian, gastric, colorectal, pancreatic, melanoma, prostate, and endometrial cancers. The Hereditary Cancer Panel (HCP) includes 76 genes. Pathogenic variants in these genes are associated with clinically actionable results which will directly impact medical management recommendations and disease risk assessment.
HCP analysis will allow the assessment of an individual’s genetic test results which, in combination with their personal and family cancer history, will assist the clinician in determining an optimum pathway for their patient’s immediate medical and/or surgical management along with clinical follow up.
Test Panels:
Hereditary Comprehensive Cancer Panel: AIP, APC, ATM, AXIN2, BAP1, BARD1, BMPR1A, BRCA1, BRCA2, BRIP1, CDC73, CDH1, CDK4, CDKN1B, CDKN2A, CHEK2, CTNNA1, DICER1, EGFR, EGLN1, EPCAM, EXT1, EXT2, FH, FLCN, GALNT12, GREM1, HOXB13, KIT, LZTR1, MAX, MEN1, MET, MITF, MLH1, MLH3, MSH2, MSH3, MSH6, MUTYH, NBN, NF1, NF2, NTHL1, PALB2, PDGFRA, PMS2, POLD1, POLE, POT1, PRKAR1A, PTCH1, PTEN, RAD51C, RAD51D, RB1, RECQL, RET, RNF43, RPS20, SDHA, SDHAF2, SDHB, SDHC, SDHD, SMAD4, SMARCA4, SMARCB1, SMARCE1, STK11, SUFU, TMEM127, TP53, TSC1, TSC2, VHL
Hereditary Breast/Ovarian/Prostate Panel: ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EPCAM, HOXB13, MLH1, MSH2, MSH6, PALB2, PMS2, PTEN, RAD51C, RAD51D, STK11, TP53
Hereditary Endometrial Panel: BRCA1, BRCA2, EPCAM, MLH1, MSH2, MSH6, PMS2, POLD1, POLE, PTEN
Hereditary Gastrointestinal Panel: APC, ATM, BMPR1A, BRCA1, BRCA2, CDH1, CDKN2A, CHEK2, CTNNA1, EPCAM, GREM1, MLH1, MLH3, MSH2, MSH3, MSH6, MUTYH, NTHL1, PALB2, PMS2, POLD1, POLE, PTEN, SDHB, SDHD, SMAD4, STK11, TP53
Hereditary Lynch Syndrome Panel: EPCAM, MLH1, MSH2, MSH6, PMS2
Hereditary Gastric Panel: APC, ATM, BRCA1, BRCA2, CDH1, CTNNA1, EPCAM, MLH1, MSH2, MSH6, PALB2, PMS2, SDHB, SDHD, SMAD4, STK11, TP53
Hereditary Pancreatic Panel: ATM, BRCA1, BRCA2, CDKN2A, EPCAM, MLH1, MSH2, MSH6, PALB2, PMS2, STK11, TP53
Hereditary Polyposis Panel: APC, BMPR1A, EPCAM, GREM1, MLH1, MLH3, MSH2, MSH3, MSH6, MUTYH, NTHL1, PMS2, POLD1, POLE, PTEN, SMAD4, STK11, TP53
Familial Gastrointestinal Stromal Panel: KIT, PDGFRA, SDHA, SDHAF2, SDHB, SDHC, SDHD
Familial Melanoma Panel: BAP1, BRCA2, CDK4, CDKN2A, MITF, POT1, PTEN
Familial Renal Panel: BAP1, FH, FLCN, MET, MITF, PTEN, SDHA, SDHAF2, SDHB, SDHC, SDHD, TP53, TSC1, TSC2, VHL
Hereditary Pheochromocytoma/Paraganglioma Panel: FH, MAX, MEN1, NF1, RET, SDHA, SDHAF2, SDHB, SDHC, SDHD, TMEM127, VHL
Central Nervous System Cancer Panel: APC, EPCAM, LZTR1, MLH1, MSH2, MSH6, NF1, NF2, PMS2, POLE, POT1, PTCH1, PTEN, SMARCB1, SMARCE1, SUFU, TP53, TSC1, TSC2, VHL
Soft Tissue Cancer Panel: APC, ATM, BRCA1, BRCA2, CHEK2, EPCAM, MLH1, MSH2, MSH6, NF1, PMS2, TP53
Please see requisition for additional small panel orderables.
Test Indications:
Cancers suspected of being of hereditary in origin frequently bear a number of hallmarks: multiple cancers in one individual or in multiple family members, younger age at diagnosis and rare or specific cancer types. Testing strategy should follow the guidelines provided by Cancer Care Ontario.
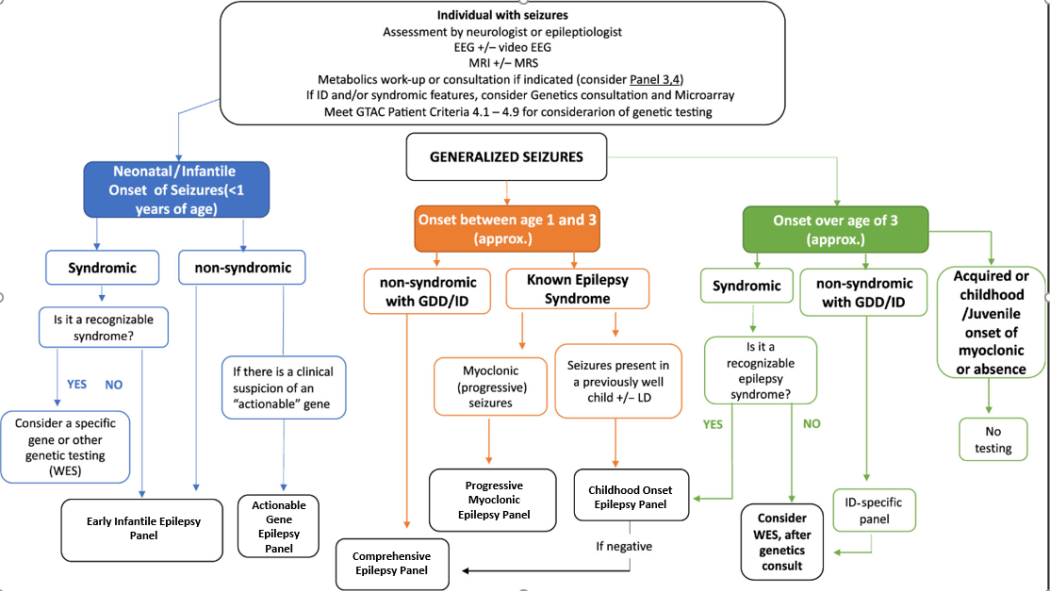
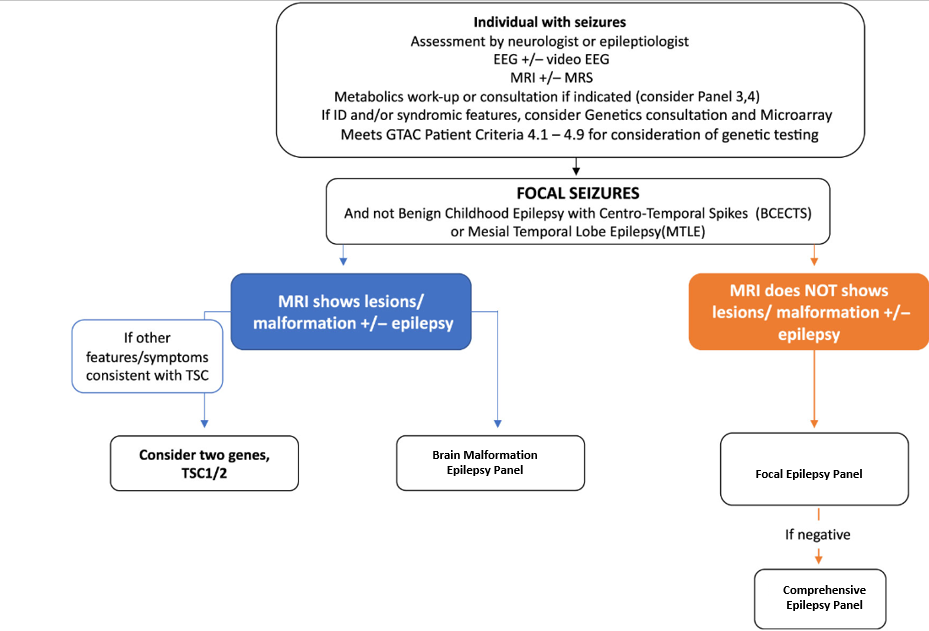
Epilepsy Panels
Test Description:
Epilepsy is a clinically heterogeneous disease with diverse aetiologies. Advances in molecular genetics over the last ten years have led to an explosion of novel genes implicated in monogenic and complex genetic epilepsies. Therefore, genetic testing now has become a critical part of the diagnostic evaluation of adults and children with epilepsy to identify genetic epilepsy syndromes, guide treatment, optimize genetic counseling, and bring closure and peace of mind to the families of those with a genetic disease whether treatable or not.
Based on the recommendations of an MOH epilepsy genetics advisory group, the Ontario Epilepsy Genetics Testing Program [OEGTP] provides evidence-based epilepsy gene panels (PMID:31587668) for properly selected epilepsy patients. The latest revision of panel genes was performed in December 2023.
The test is designed to provide rule out level assessment for all coding sequence and copy number alterations for 190 genes, which encompass the majority of the known genetic etiologies for epilepsy. The test includes all major epilepsy syndromes and is subdivided into seven clinical categories:
- Comprehensive Epilepsy Panel (190 genes)
- Focal Epilepsy Panel (12 genes)
- Progressive Myoclonic Epilepsy Panel (21 genes)
- Early Infantile Epilepsy Panel (84 genes)
- Childhood Onset Epilepsy Panel (59 genes)
- Brain Malformation Epilepsy Panel (45 genes)
- Actionable Gene Epilepsy Panel (25 genes)
Please note that both sequence and copy number alterations (deletions/duplications) are routinely tested for every gene.
Test Panels:
Epilepsy Comprehensive panel: 190 Genes
ABAT, ACTB, ACTG1, ADGRG1, ADSL, AFG2A, AKT3, ALDH7A1, ALG13, AMT, AP3B2, ARFGEF2, ARHGEF9, ARV1, ARX, ASAH1, ASNS, ATP1A2, ATP1A3, ATP6V0A2, ATP7A, ATRX, B3GALNT2, CACNA1A, CACNA1E, CAD, CDKL5, CHD2, CHRNA4, CHRNB2, CLCN4, CLN3, CLN5, CLN6, CLN8, CNTNAP2, CSTB, CTSD, CTSF, DCX, DEPDC5, DNAJC5, DNM1, DOCK7, DYNC1H1, DYRK1A, EEF1A2, EHMT1, EPM2A, FGF12, FKRP, FKTN, FLNA, FOLR1, FOXG1, FRRS1L, GABBR2, GABRA1, GABRB2, GABRB3, GABRG2, GAMT, GLDC, GMPPB, GNAO1, GOSR2, GPSM2, GRIN1, GRIN2A, GRIN2B, GRIN2D, GRN, HCN1, HNRNPU, ITPA, KANSL1, KATNB1, KCNA1, KCNA2, KCNB1, KCNC1, KCNH5, KCNJ10, KCNMA1, KCNQ2, KCNQ3, KCNT1, KCTD7,KDM5C, KIF2A, LAMA2, LARGE1, LGI1, MBD5, MDH2, MECP2, MEF2C, MFSD8, MOCS1, NDE1, NEU1, NEXMIF, NGLY1, NHLRC1, NPRL2, NPRL3, NRXN1, OCLN, PAFAH1B1, PAK3, PCDH19, PHF6, PHGDH, PIGA, PIGG, PIGN, PIGO, PIGT, PIGV, PLCB1, PLPBP, PNKP, PNPO, POLG, POMGNT1, POMGNT2, POMK, POMT1, POMT2, PPT1, PRRT2, PSAT1, PSPH, PURA, RAB18, RAB39B, RAB3GAP1, RAB3GAP2, RELN, ROGDI, RTTN, SCARB2, SCN1A, SCN1B, SCN2A, SCN3A, SCN8A, SERPINI1, SGCE, SLC12A5, SLC13A5, SLC19A3, SLC25A12, SLC25A22, SLC2A1, SLC35A2, SLC6A1, SLC6A8, SLC9A6, SMARCA2, SNAP29, SPTAN1, SRD5A3, ST3GAL5, STX1B, STXBP1, SUOX, SYN1, SYNGAP1, SYNJ1, SZT2, TBC1D24, TCF4, TPP1, TRPM3, TSC1, TSC2, TUBA1A, TUBB, TUBB2A, TUBB2B, TUBB3, UBA5, UBE3A, VLDLR, WDR45, WDR62, WWOX, YWHAG, ZEB2
Focal Epilepsy panel: 12 Genes
CHRNA4, CHRNB2, DEPDC5, GRIN2A, KCNT1, LGI1, NPRL2, NPRL3, PRRT2, SCN1A, SCN1B, SLC2A1
Progressive Myoclonic Epilepsy panel: 21 Genes
ASAH1, CLN3, CLN5, CLN6, CLN8, CSTB, CTSD, CTSF, EPM2A, GOSR2, GRN, KCNC1, KCTD7, MFSD8, NEU1, NHLRC1, PPT1, SCARB2, SERPINI1, SGCE, TPP1
Early Infantile Epilepsy panel: 84 Genes
ABAT, ADSL, AFG2A, ALDH7A1, ALG13, AP3B2, ARHGEF9, ARV1, ARX, CACNA1A, CACNA1E, CAD, CDKL5, CHD2, DCX, DNM1, DOCK7, DYRK1A, EEF1A2, FGF12, FOLR1, FOXG1, FRRS1L, GABBR2, GABRA1, GABRB2, GABRB3, GABRG2, GAMT, GLDC, GNAO1, GRIN2A, GRIN2B, GRIN2D, HCN1, HNRNPU, ITPA, KCNA1, KCNA2, KCNB1, KCNH5, KCNQ2, KCNQ3, KCNT1, MDH2, MECP2, MEF2C, NGLY1, PCDH19, PIGA, PIGG, PIGN, PIGO, PIGT, PIGV, PLCB1, PNKP, PNPO, POLG, PRRT2, PURA, SCN1A, SCN1B, SCN2A,SCN8A, SLC12A5, SLC13A5, SLC25A12, SLC25A22, SLC2A1, SLC35A2, SLC6A8, SPTAN1, ST3GAL5, STX1B, STXBP1, SYNGAP1, SYNJ1, SZT2, TBC1D24, UBA5, WDR45, WWOX, YWHAG
Childhood Onset Epilepsy panel: 59 Genes
ADSL, ARX, ATP1A3, ATRX, CDKL5, CHD2, CLCN4, CNTNAP2, DEPDC5, DNAJC5, DYRK1A, EHMT1, FOXG1, GABBR2, GABRB2, GABRG2,GRIN2A, GRIN2D, KANSL1, KCNJ10, KCNMA1, KCNQ3, KDM5C, MBD5, MECP2, MEF2C, NEXMIF, NGLY1, NRXN1, PAK3, PCDH19, PHF6, PIGA, PIGN, PIGO, PNKP, POLG, PRRT2, RAB39B, ROGDI, SCN1A, SCN1B, SCN2A, SLC2A1, SLC6A1,SLC6A8, SLC9A6, SMARCA2, STX1B, SYN1, SYNGAP1, TBC1D24, TCF4, TRPM3, TSC1, TSC2, UBE3A, WDR45, ZEB2
Brain Malformation Epilepsy panel: 45 Genes
ACTB, ACTG1, ADGRG1, AKT3, ARFGEF2, ARX, ASNS, ATP6V0A2, B3GALNT2, B3GNT1(B4GAT1), DCX, DYNC1H1, FKRP, FKTN, FLNA, GMPPB, GPR56(ADGRG1), GPSM2, GTDC2(POMGNT2), KATNB1, KIAA1279(KIF1BP), KIF2A, LAMA2, LARGE, NDE1, OCLN, PAFAH1B1, POMGNT1, POMT1, POMT2, RAB18, RAB3GAP1, RAB3GAP2, RELN, RTTN, SGK196(POMK), SNAP29, SRD5A3, TUBA1A, TUBB, TUBB2A, TUBB2B, TUBB3, VLDLR, WDR62
Actionable Gene Epilepsy panel: 25 Genes
ALDH7A1, AMT, ATP7A, CAD, FOLR1, GAMT, GLDC, KCNQ2, KCNT1, MOCS1, PHGDH, PLPBP, PNPO, POLG, PSAT1, PSPH, SCN1A, SLC19A3, SLC2A1, SLC6A8, SUOX, TPP1, TRPM3, TSC1, TSC2
Test Indications
This Epilepsy panel test is a deep sequencing NGS assay designed as a rule out sequencing and copy number analysis test for all coding sequences of all genes tested. Content is designed by a panel of clinical experts Ontario MOHLTC Genetic Epilepsy Working Group to include majority of genes associated with epilepsy as the cardinal clinical presentation. In patients where epilepsy is not the cardinal clinical feature, and genetic etiology is suspected, other genetic and genomic analyses and clinical genetics referral may be considered.
Ontario Physicians must include completed questionnaire prior to submitting specimens to PaLM and confirm that age of onset, seizure type and electroclinical syndrome is consistent with a genetic epilepsy. Epilepsy Genetic Test Requistion
The etiologies of genetic epilepsies are heterogeneous and a significant proportion of cases are attributable to structural brain defects and inherited metabolic disorders. Often it can be difficult to predict genotype based on electro-clinical phenotype. Individuals with epilepsy who may benefit from genetic testing include those with infantile onset, epilepsy refractory to treatment, epilepsy plus developmental delay, or families that may choose to have prenatal testing in future pregnancy. As this is an evolving field, the genotype-phenotype correlations are not well understood, and there is considerable phenotypic variability within the same genetic defect. For instance, mutations in the sodium channel genes cause Dravet syndrome as well as generalized epilepsy with febrile seizure plus. A particular electroclinical syndrome may be caused by multiple genetic defects. For example, Ohtahara syndrome may be caused by mutations in syntaxin binding protein-1 (STXBP1), potassium channel mutations (KCNQ2), Aristaless related homeobox (ARX), and solute carrier family 25, member 22 (SLC25A22) encoding a mitochondrial glutamate carrier.
Familial Dyslipidemia/Hypercholesterolemia Panels
Test Description:
Abnormal lipid levels (dyslipidemias) are the major drivers of atherosclerosis (AS), one of the major risk factors for cardiovascular disease (CVD). Familial hypercholesterolemia (FH) is one of the most common monogenic disorders, affecting approximately 1 in 250-300 Canadians, yet is underdiagnosed in Canada and worldwide. DNA-based test is a component of the diagnostic algorithms for dyslipidemias as described by the Simon Broome Register criteria (Marks et al., 2003) the Dutch Lipid Clinic Network criteria (Fouchier et al., 2001) and the Canadian Cardiovascular Society (Ruel et al., 2018). This Familial Hypercholesterolemia/Dyslipidemia panel test is a deep sequencing NGS assay designed based on guidance from the Ontario Health-FH Working Group and clinical specialists.
Please note that this test cannot be ordered for Alzheimer’s disease/dementia.
Test Panels:
Familial Hypercholesterolemia Panel (8): ABCG5, ABCG8, APOB, APOE, LDLR, LDLRAP1, LIPA, PCSK9
Dyslipidemia – Comprehensive Panel (25): ABCA1, ABCG5, ABCG8, ANGPTL3, APOA1, APOA5, APOB, APOC2, APOC3, APOE, CETP, GPD1, GPIHBP1, LCAT, LDLR, LDLRAP1, LIPA, LIPC, LMF1, LPL, MTTP, PCSK9, SAR1B, SCARB1, STAP1
Please note that the Comprehensive Dyslipidemia Panel is only available to providers outside of Ontario.
Test Indications:
Individuals being tested should meet one or more of the following:
- Confirmed FH disease-causing pathogenic/likely pathogenic variant in a close blood relative
- High LDL-cholesterol level (not due to secondary causes
- Untreated LDL-cholesterol level ≥5.0 mmol/L for age 40years and over
- Untreated LDL-cholesterol level ≥4.5 mmol/L for age 18 to 39 years
- Untreated LDL-cholesterol level ≥3.5 mmol/L for age under 18 years
AND at least one of the following
- Tendon xanthomas and/or corneal arcus in proband
- First-degree relative (FDR) with high LDL-cholesterol level not due to secondary causes
- Proband or FDR with early onset ASCVD (<55years for men; <65years for women)
- Limited family history information (eg individual is adopted)
Charcot-Marie-Tooth Gene Panel
Test Description:
Charcot-Marie-Tooth (CMT) disease is a genetically and clinically heterogeneous group of inherited disorders of the peripheral nervous system characterized by a chronic motor and sensory polyneuropathy, also known as hereditary motor and sensory neuropathy (HMSN). CMT was previously classified by type: demyelinating (CMT1), Axonal or non-demyelinating (CMT2), and Dominant Intermediate (DI-CMT). A newly proposed naming system incorporates the gene in recognition of phenotypic overlap between types. The most common pathogenic variant is associated with CMT1A (70%-80% of all CMT1) and involves duplication of PMP22, while PMP22 gene deletion is the most common cause (80%) of hereditary neuropathy with liability to pressure palsies (HNPP).
Mutations in remaining genes are associated with less frequent subtypes of CMT, including autosomal dominant, recessive and X-linked forms of the disease.
Test Panels:
Charcot-Marie-Tooth panel-Comprehensive (87): AARS, ABHD12, AIFM1, ARHGEF10, ARHGEF28, ATP1A1, ATP7A, BAG3, BSCL2, C1orf194, CNTNAP1, DCTN1, DCTN2, DGAT2, DHTKD1, DNAJB2, DNM2, DNMT1, DRP2, DYNC1H1, EGR2, FBLN5, FGD4, FIG4, GARS, GDAP1, GJB1, GNB4, HARS, HINT1, HSPB1, HSPB3, HSPB8, IGHMBP2, INF2, JAG1, KARS, KIF1B, KIF5A, LITAF, LMNA, LRSAM1, MARS, MCM3AP, MFN2, MME, MORC2, MPV17, MPZ, MTMR2, NAGLU, NDRG1, NEFH, NEFL, PDK3, PDXK, PLEKHG5, PMP2, PMP22, PNKP, PRPS1, PRX, PTRH2, RAB7A, SBF1, SBF2, SCO2, SELRC1, SEPT9, SETX, SGPL1, SH3TC2, SIGMAR1, SLC12A6, SLC9A3R1, SORD, SPG11, SPTLC1, SURF1, TFG, TRIM2, TRPV4, TTR, VCP, VRK1, WARS, YARS
Test Indications:
This test is intended for use as a diagnostic aide for individuals with clinical suspicion of Charcot-Marie-Tooth hereditary neuropathy (CMT) or Hereditary Neuropathy with Liability to Pressure Palsies (HNPP) or for individuals with a known family history of these conditions and a known genetic variation.
Mitochondrial Gene Panel
Test Description:
The human mitochondrial DNA (mtDNA) encodes 37 genes coding for two rRNAs, 22 tRNAs and 13 polypeptides within its 16 569 bp. The mtDNA-encoded polypeptides are all subunits of enzyme complexes of the oxidative phosphorylation system. Disease phenotypes resulting from mitochondrial mutations may appear as distinct syndromes, such as Kearns-Sayre syndrome (KSS), Leber’s Hereditary Optic Neuropathy (LHON), mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes (MELAS), chronic progressive external ophthalmoplegia (CPEO), myoclonic epilepsy with ragged-red fibers (MERRF), neurogenic weakness with ataxia and retinitis pigmentosa (NARP) or Leigh syndrome (LS). More frequently, the clinical presentation is much more heterogeneous. Some common symptoms include ptosis, external ophthalmoplegia, proximal myopathy, exercise intolerance, cardiomyopathy, sensorineural deafness, migraine, stroke-like episodes, pigmentary retinopathy, diabetes mellitus, encephalopathy, seizures, ataxia, and spasticity. This panel has been augmented with a selected series of 19 nuclear genes known to be associated the mitochondrial depletion disorders. About 80-95% of patients with mitochondrial disorders do not harbor a pathogenic mutation in the mitochondrial genome. A large proportion of these cases may have defects in nuclear-encoded genes that are involved in the biosynthesis of the mitochondrial genome or in the maintenance of mtDNA integrity. The Mitochondrial Genome Sequencing and Depletion/Integrity panel is appropriate for patients suspected of having one of the various forms of mtDNA depletion syndrome and/or mtDNA multiple deletions.
Mitochondrial and Nuclear genes are a single panel
Mitochondrial Encoded:
MT-TY, MT-TW, MT-TV, MT-TT, MT-TS2, MT-TS1, MT-TR, MT-TQ, MT-TP, MT-TN, MT-TM, MT-TL2, MT-TL1, MT-TK, MT-TI, MT-TH, MT-TG, MT-TF, MT-TE, MT-TD, MT-TA, MT-RNR2, MT-RNR1, MT-ND6, MT-ND5, MT-ND4L ,MT-ND4 ,MT-ND3 ,MT-ND2 ,MT-ND1, MT-CYB, MT-CO3, MT-CO2, MT-CO1, MT-TC, MT-ATP8, MT-ATP6
Nuclear Encoded:
APTX, DGUOK, DNA2, FBXL4, GFER, MGME1, MPV17, OPA1, OPA3(isoformA & B), POLG, POLG2, RRM2B, SLC25A4, SPG7(isoform1 & 2), SUCLA2, SUCLG1, TK2, TWNK(C10orf2), TYMP
Urea Cycle Disorders
Test Description:
Urea cycle disorders are a family of disorders in which excess ammonia found in the plasma is the result of either failure of ammonia detoxification or overproduction of ammonia. Defects in the urea cycle or its cofactors are not evident at birth, but symptoms frequently appear within days of birth. Including Cerebral edema, poor feeding and vomiting, seizures, hypothermia as well as hyper or hypoventilation. Primary hyperammonemia may be the result of an inherited deficiency of one of the enzymes composing the hepatic urea cycle and membrane transporters. Secondary hyperammonemia is the result of defects in the related pathways responsible for the assembly of ammonia acceptors and associated substrates required in the function of the urea cycle.
Heterogeneity in urea cycle disorders supports the use of a multi-gene panel for genetic testing. London Health Sciences Centre Urea Cycle Panel (LHSC UCD) is a Next Generation Sequencing (NGS) test involving sequence and copy number analysis of coding regions and adjacent intronic regions for 13 genes associated with various forms of urea cycle disorders.
The test is designed to provide rule out level assessment for all coding sequence and copy number alterations for 13 genes, which encompass the majority of the known genetic etiologies for urea cycle disorders. This test is useful in the confirmation of a clinical diagnosis, prenatal diagnosis in the presence of a family history of urea cycle disorders, as well as testing individuals with idiopathic hyperammonemia or suspicion of a urea cycle disorder.
ARG1, ASL, CA5A, SLC25A13, ASS1, GLUL, CPS1, SLC25A15, SLC25A2, GLUD1, SLC7A7, OTC, CPS1
Test Indications:
This test is useful in the confirmation of a clinical diagnosis of urea cycle disorders, prenatal diagnosis in the presence of a family history of urea cycle disorders, as well as testing individuals with idiopathic hyperammonemia or suspicion of a urea cycle disorder. This information may also be helpful when pursuing genetic counselling for at risk family members. Individual genes may be selected when a known pathogenic variant previously identified in a family member can be used to determine the status of at risk pregnancies or individuals.
Lysosomal Storage Disorders
Test Description:
Lysosomal storage disorders are a family of disorders in which there is a pathological accumulation of macromolecules in the lysosomes. This may be the result of defects in lysosomal enzyme function, or failure to transport across the lysosomal membrane. The accumulation of these macromolecules in turn result in cell damage and ultimately impaired organ function.
A list of more than 50 different inherited metabolic disorders fall under the category of lysosomal storage disorders.
The London Health Sciences Centre Lysosomal Storage Panel (LHSC LSD) is a Next Generation Sequencing (NGS) test involving sequence and copy number analysis of coding regions and adjacent intronic regions for 50 genes associated with various forms of lysosomal storage disorders. The test is designed to provide rule out level assessment for all coding sequence and copy number alterations for 50 genes, which encompass the majority of the known genetic etiologies for lysosomal storage disorders.
AGA, ARSA, ARSB, ASAH1, CLN3, CLN5, CLN6, CLN8, CTNS, CTSA, CTSD, CTSK, DNAJC5, FUCA1, GAA, GALC, GALNS, GBA, GLA, GLB1, GM2A, GNPTAB, GNPTG, GNS, GRN, GUSB, HEXA, HEXB, HGSNAT, HYAL1, IDS, IDUA, LAMP2, LIPA, MAN2B1, MANBA, MCOLN1, MFSD8, NAGA, NAGLU, NEU1, NPC1, NPC2, PPT1, PSAP, SGSH, SLC17A5, SMPD1, SUMF1, TPP1
Test Indications:
This test is useful in the confirmation of a clinical diagnosis in conjunction with biochemical analysis and postnatal and prenatal diagnosis in the presence of a family history of lysosomal storage disorders.
Atypical Hemolytic Uremic Syndrome Panel (Research Only)
Test Description:
Atypical hemolytic-uremic syndrome is a disease that primarily affects kidney function. This condition, which can occur at any age, causes thrombosis to form in small blood vessels in the kidneys. These clots can cause serious medical problems if they restrict or block blood flow. Atypical hemolytic-uremic syndrome is characterized by three major features: hemolytic anemia, thrombocytopenia, and kidney failure. Atypical hemolytic-uremic syndrome often results from a combination of environmental and genetic factors. Pathogenic variants in a number of genes can increase the risk of developing the disorder. PMID: 23251215; 24029428; 30294946
Adult Onset Hemolytic Uremic Syndrome Gene Panel
C3, C9, CD46, CFB, CFH, CFI, DGKE, F12, FKRP, INF2, MMACHC, MMADHC, PLG, THBD, VTN, VWF
This test is currently available through research use only. Funding is provided through Alexion Pharmaceuticals.
Hematologic Oncology Molecular Testing
Test Description:
Hematologic Oncology NGS 50 gene (DNA) and 674 RNA (gene fusions)
Diagnostic assessment of patients with suspected hematologic malignancies follows a complex triaged protocol involving flow cytometric analysis, anatomic pathology, cytogenetics and molecular diagnostics. Classically, molecular genetics assessment is reserved for patients for whom initial assessment by flow cytometry, cytogentics or pathology indicates a possibility of a specific molecular subtype, followed by a targeted confirmatory molecular assay. This approach suffers from limitations including: inefficient coordination of complex triage procedures between different laboratories; insufficient specimens (often bone marrow) for repeat testing; increased turnaround times, and low diagnostic yield. In routine clinical use, a NGS-based sequencing and gene fusion panel will be utilized for every patient specimen, in parallel to the standard karyotype assessment and flow cytometry. The panel includes assessment of 50 key DNA target genes, along with 30 driver genes involved in over 600 clinically-relevant gene fusions. Clinical validations highlighted an analytical sensitivity of 5% for detection of DNA sequence mutations (including small in/dels and more complex mutation such as FLT3 ITD) and 1% for detection of gene fusions. This approach offers considerably simplified molecular testing protocol (single common assay for all specimens), reduced TATs, and substantially increased molecular diagnostic yield in this patient population.
Test Panels:
DNA Mutation Hotspots sequencing (28): ABL1, ANKRD26, BRAF, CBL, CSF3R, DDX41, DNMT3A, FLT3, GATA2, HRAS, IDH1, IDH2, JAK2, KIT, KRAS, MPL, MYD88, NPM1, NRAS, PPM1D, PTPN11, SETBP1, SF3B1, SMC1A, SMC3, SRSF2, U2AF1, WT1
Full genes sequencing (23): ASXL1, BCOR, BCORL1, CALR, CEBPA, CUX1, ETV6, EZH2, IKZF1, NF1, PHF6, PRPF8, RB1, RUNX1, SH2B3, STAG2, TERC, TERT, TET2, TP53, ZRSR2
Gene fusions: ABL1, ALK, BCL2, BRAF, CCND1, CREBBP, CRE, BBP, EGFR, ETV6, FGFR1, FGFR2, FUS, HMGA2, JAK2, KMT2A (MLL-PTD), MECOM, MET, MLLT10, MLLT3, MYBL1, MYH11, NTRK3, NUP214, PDGFRA, PDGFRB, RARA, RBM15, RUNX1, TCF3, TFE3
Note: Each of the above “driver” fusion genes may have multiple fusion partners. This assay includes 674 of the most common fusion partner combinations of the above listed genes. For details please refer to: Thermo Fisher Ion AmpliSeq™ Oncomine Myeloid Assay.
Targeted Myeloid DNA Assays:
- FLT3 D835 TKD & ITD
- JAK2 (p.V617F)
- NPM1 (p.W288fs*12)
Targeted Myeloid Fusion (RNA) Assays: (for disease monitoring only)
- BCR::ABL1 p210 t(9;22) fusion (CML)
- BCR::ABL1 p190 t(9;22) fusion
- CBFB::MYH11 Type A t(16;16)/inv(16) fusion
- PML::RARA t(15;17) fusion: bcr1, bcr2, bcr3
- RUNX1::RUNX1T1 t(8;21) fusion
For providers outside of LHSC: please note that this testing is billed to the ordering provider. Please contact the laboratory genetic counsellor at lisa.karger@lhsc.on.ca or 519-685-8500, Extension. 77083 for guidance.
Targeted Gene Testing
Test Description:
Inherited disorders are frequently the result of predominantly single gene defects (for example, CADASIL as the result of NOTCH3 pathogenic variants, Medium Chain Acyl CoA Dehydrogenase (MCAD) Deficiency as a result of pathogenic variants in ACADM pathogenic variants or Non-Syndromic Autosomal Recessive Deafness as a result of pathogenic variants in GJB2 and GJB6). Analysis of these genes can be ordered using the General and Biochemistry Requisitions.
A select group of diseases are overwhelmingly represented by a short list of specific variants, as in Cystic Fibrosis, Factor V Leiden, HFE-related haemochromatosis, DPYD and Prothrombin.
See Test Menu for specific gene information.


